Volume 35, Issue 246 (7-2025)
J Mazandaran Univ Med Sci 2025, 35(246): 60-71 |
Back to browse issues page
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
Imani Baran A, Alidoost S, Nematollahi A, Firouzamandi M, Ghorbanzadeh B. Identification of Dicrocoelium dendriticum isolated from small ruminants of east Azerbaijan province using ITS2 and Nad1 gene markers. J Mazandaran Univ Med Sci 2025; 35 (246) :60-71
URL: http://jmums.mazums.ac.ir/article-1-21251-en.html
URL: http://jmums.mazums.ac.ir/article-1-21251-en.html
Abbas Imani Baran 
 , Salar Alidoost
, Salar Alidoost 
 , Ahmad Nematollahi
, Ahmad Nematollahi 
 , Masoomeh Firouzamandi
, Masoomeh Firouzamandi 
 , Behzad Ghorbanzadeh
, Behzad Ghorbanzadeh 


 , Salar Alidoost
, Salar Alidoost 
 , Ahmad Nematollahi
, Ahmad Nematollahi 
 , Masoomeh Firouzamandi
, Masoomeh Firouzamandi 
 , Behzad Ghorbanzadeh
, Behzad Ghorbanzadeh 

Abstract: (819 Views)
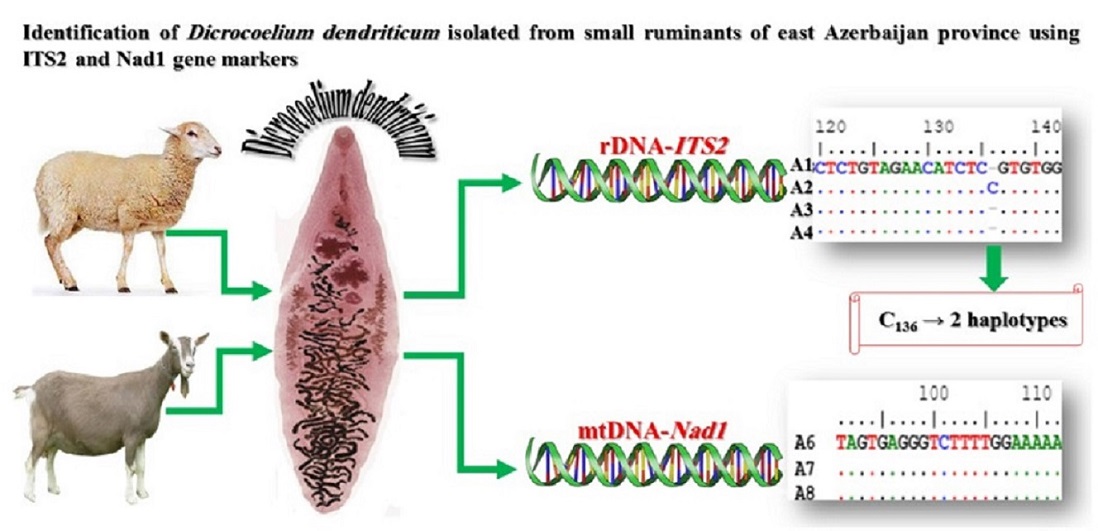
Background and purpose: Dicrocoelium dendriticum is the most important liver fluke of the domestic and wild herbivores. Nevertheless, there is scarce data on the genetic characteristics of this important zoonotic trematode. This study aimed to determine the intraspecific variations within D. dendriticum samples from sheep and goats in East Azerbaijan province, northwestern Iran, through the analysis of mitochondrial Nad1 and ribosomal ITS2 genes.
Materials and methods: In this study, DNA was extracted from the D. dendriticum samples. Subsequently, the Nad1 and ITS2 genes were amplified through conventional PCR method. PCR products from each gene for sheep (6 samples) and goat (4 samples) isolates were sent for sequencing. The obtained sequences were edited through BioEdit software and aligned with each other using the ClustalW algorithm in MEGA7.0 software. Similarly, all nucleotide sequences were compared with some sequences deposited in the NCBI, based on BLAST function. Finally, to determine the phylogenetic status of D. dicrocoelium isolates, a phylogenetic tree relevant to each gene marker was constructed using the Neighbor-Joining method in MEGA7.0 software to investigate the similarities and differences between isolates from different geographical regions around the world and our D. dendriticum samples.
Results: The analysis of sequences showed that, among the 4 ITS2 sequences, there was only one additional nucleotide relevant to one of the sheep isolates, while no difference was found among the sequences of Nad1 fragments. In the BLAST comparison, except for a few sequences, 100% similarity was found between our sequences and reference sequences from NCBI. Phylogenetic tree construction indicated that both ITS2 sequences and Nad1 fragments were clearly grouped in a clade, both separately and together, with the majority of selected sequences from NCBI.
Conclusion: The haplotype difference was not observed for the Nad1 gene, so it is better to use the ITS2 gene to detect different haplotypes of D. dendriticum.
Materials and methods: In this study, DNA was extracted from the D. dendriticum samples. Subsequently, the Nad1 and ITS2 genes were amplified through conventional PCR method. PCR products from each gene for sheep (6 samples) and goat (4 samples) isolates were sent for sequencing. The obtained sequences were edited through BioEdit software and aligned with each other using the ClustalW algorithm in MEGA7.0 software. Similarly, all nucleotide sequences were compared with some sequences deposited in the NCBI, based on BLAST function. Finally, to determine the phylogenetic status of D. dicrocoelium isolates, a phylogenetic tree relevant to each gene marker was constructed using the Neighbor-Joining method in MEGA7.0 software to investigate the similarities and differences between isolates from different geographical regions around the world and our D. dendriticum samples.
Results: The analysis of sequences showed that, among the 4 ITS2 sequences, there was only one additional nucleotide relevant to one of the sheep isolates, while no difference was found among the sequences of Nad1 fragments. In the BLAST comparison, except for a few sequences, 100% similarity was found between our sequences and reference sequences from NCBI. Phylogenetic tree construction indicated that both ITS2 sequences and Nad1 fragments were clearly grouped in a clade, both separately and together, with the majority of selected sequences from NCBI.
Conclusion: The haplotype difference was not observed for the Nad1 gene, so it is better to use the ITS2 gene to detect different haplotypes of D. dendriticum.
Type of Study: Research(Original) |
Subject:
parasitology
Send email to the article author
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |