Volume 35, Issue 247 (8-2025)
J Mazandaran Univ Med Sci 2025, 35(247): 49-60 |
Back to browse issues page
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
Nooraei F, Mohseni M. Investigating the Prevalence of the Vancomycin Resistance Gene in Staphylococcus aureus Isolated from Clinical Specimens in Medical Centers in Mazandaran. J Mazandaran Univ Med Sci 2025; 35 (247) :49-60
URL: http://jmums.mazums.ac.ir/article-1-21912-en.html
URL: http://jmums.mazums.ac.ir/article-1-21912-en.html
Abstract: (763 Views)
Background and purpose: Staphylococcus aureus is one of the most important causative agents of infections. The excessive use of antibiotics has led to a rise in antibiotic-resistant strains, including vancomycin-resistant isolates. The aim of this study was to investigate the prevalence of the vanA gene** in S. aureus strains isolated from clinical specimens.
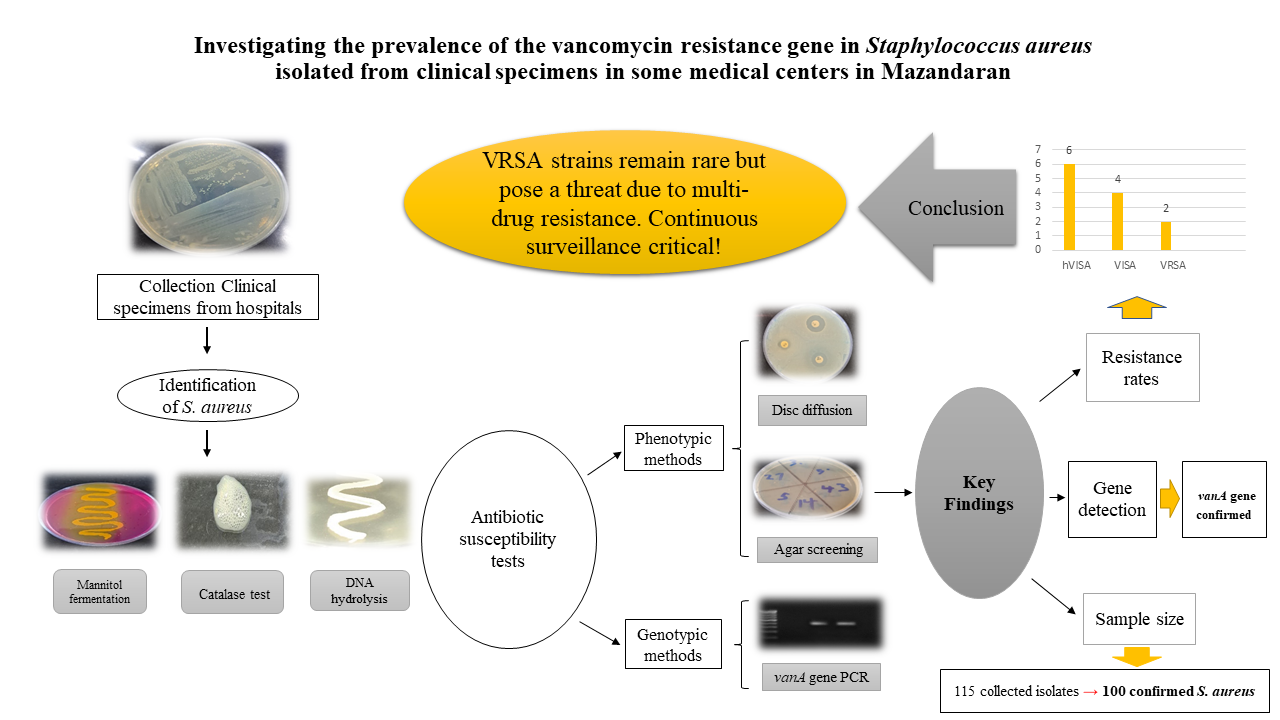
Materials and methods: A total of 115 S. aureus isolates were collected from medical diagnostic laboratories and hospitals. The isolates were identified using standard phenotypic methods. The antibiotic resistance profiles of the isolates to commonly used antibiotics were assessed using the disc diffusion method. Vancomycin susceptibility was evaluated using the vancomycin agar screening method, and the minimum inhibitory concentration (MIC) of vancomycin was determined by the agar dilution method. The presence of the vanA gene was detected using polymerase chain reaction (PCR). Statistical analysis was performed using SPSS software, employing Fisher’s exact test and the Mann–Whitney U test.
Results: A total of 100 isolates were confirmed as S. aureus. The resistance rates to penicillin, erythromycin, clindamycin, tetracycline, ciprofloxacin, cefoxitin, gentamicin, and rifampin were 90%, 66%, 56%, 54%, 54%, 38%, 4%, and 0%, respectively. The results of vancomycin susceptibility testing, using both phenotypic and genotypic methods, showed that 2% of the isolates were resistant, and 4% exhibited intermediate resistance to vancomycin.
Conclusion: The results indicate that the prevalence of resistance genes and the emergence of vancomycin-resistant S. aureus (VRSA) have not changed significantly compared to previous studies. However, given the multidrug resistance of VRSA isolates to many commonly used antibiotics, it is essential to accurately identify and effectively manage infections caused by these strains.
Materials and methods: A total of 115 S. aureus isolates were collected from medical diagnostic laboratories and hospitals. The isolates were identified using standard phenotypic methods. The antibiotic resistance profiles of the isolates to commonly used antibiotics were assessed using the disc diffusion method. Vancomycin susceptibility was evaluated using the vancomycin agar screening method, and the minimum inhibitory concentration (MIC) of vancomycin was determined by the agar dilution method. The presence of the vanA gene was detected using polymerase chain reaction (PCR). Statistical analysis was performed using SPSS software, employing Fisher’s exact test and the Mann–Whitney U test.
Results: A total of 100 isolates were confirmed as S. aureus. The resistance rates to penicillin, erythromycin, clindamycin, tetracycline, ciprofloxacin, cefoxitin, gentamicin, and rifampin were 90%, 66%, 56%, 54%, 54%, 38%, 4%, and 0%, respectively. The results of vancomycin susceptibility testing, using both phenotypic and genotypic methods, showed that 2% of the isolates were resistant, and 4% exhibited intermediate resistance to vancomycin.
Conclusion: The results indicate that the prevalence of resistance genes and the emergence of vancomycin-resistant S. aureus (VRSA) have not changed significantly compared to previous studies. However, given the multidrug resistance of VRSA isolates to many commonly used antibiotics, it is essential to accurately identify and effectively manage infections caused by these strains.
Type of Study: Research(Original) |
Subject:
Microbiology
Send email to the article author
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |